Example: iVar vs IRMA
The accuracy and completeness of iVar and IRMA assemblies were determined for 6 virus species (Alphainfluenzavirus influenzae, Betainfluenzavirus influenzae, Mobillivirus hominis, Orthorubulavirus parotitidis, Severe acute respiratory syndrome coronavirus 2, and Enterovirus D) using Illumina hybrid capture data generated from 7 Twist Bioscience synthetic DNA controls (controls 2, 4, 5, 6, 15, 17, and 66). Multiple replicates of controls 5, 15, and 17 were included. Assembly accuracy and completeness were calculated using the scripts located here.
VAPER + iVar vs VAPER + IRMA
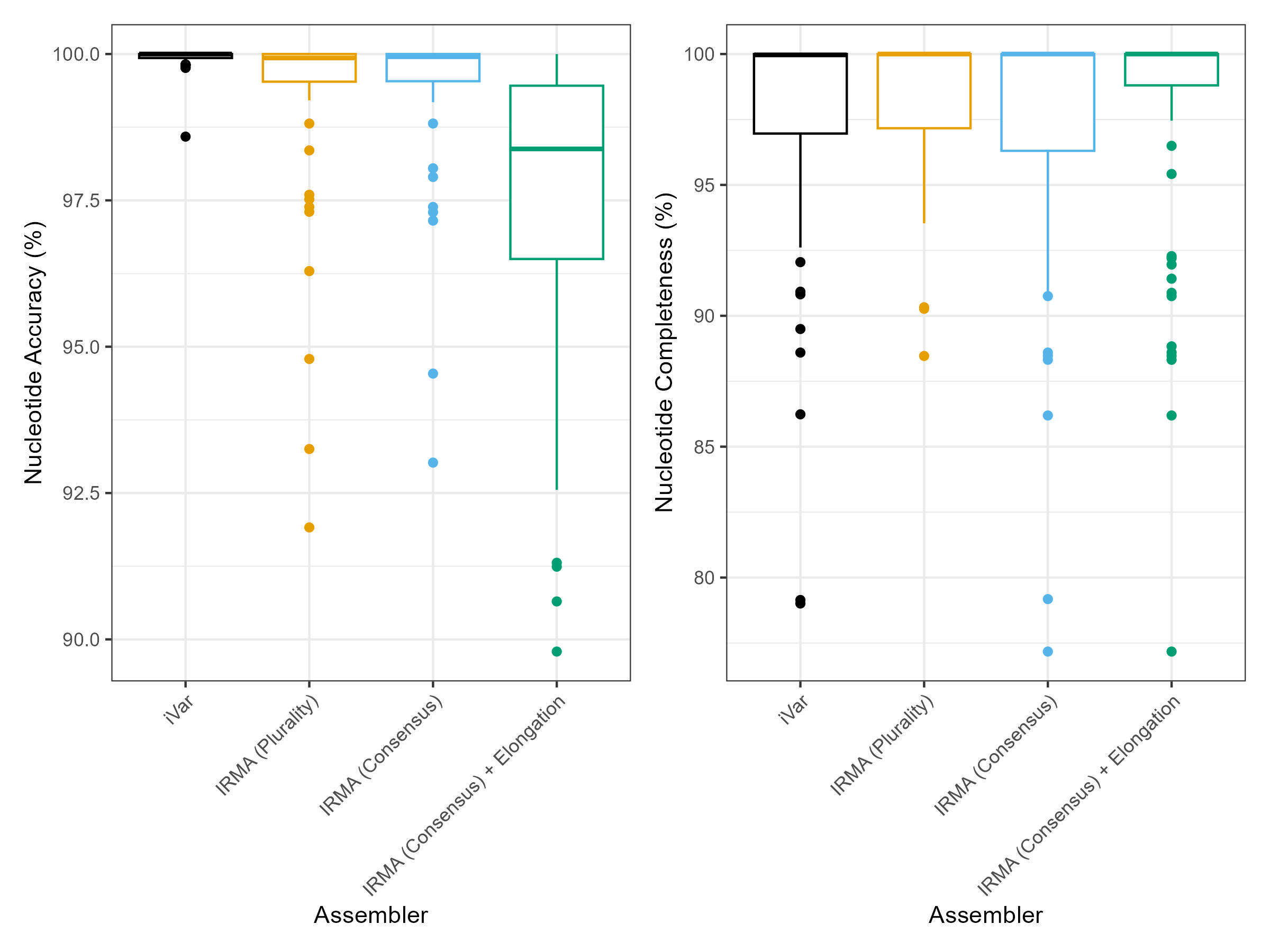
Assemblies were created from each synthetic control using VAPER with the EPITOME_2025-02-14 reference set with the following conditions:
- iVar:
--cons_assembler ivar --cons_assembly_type consensus - IRMA (Consensus):
--cons_assembler irma --cons_assembly_type consensus - IRMA (Consensus + Elongation):
--cons_assembler irma --cons_assembly_elong true - IRMA (Plurality):
--cons_assembler irma --cons_assembly_type plurality
VAPER vs IRMA FLU
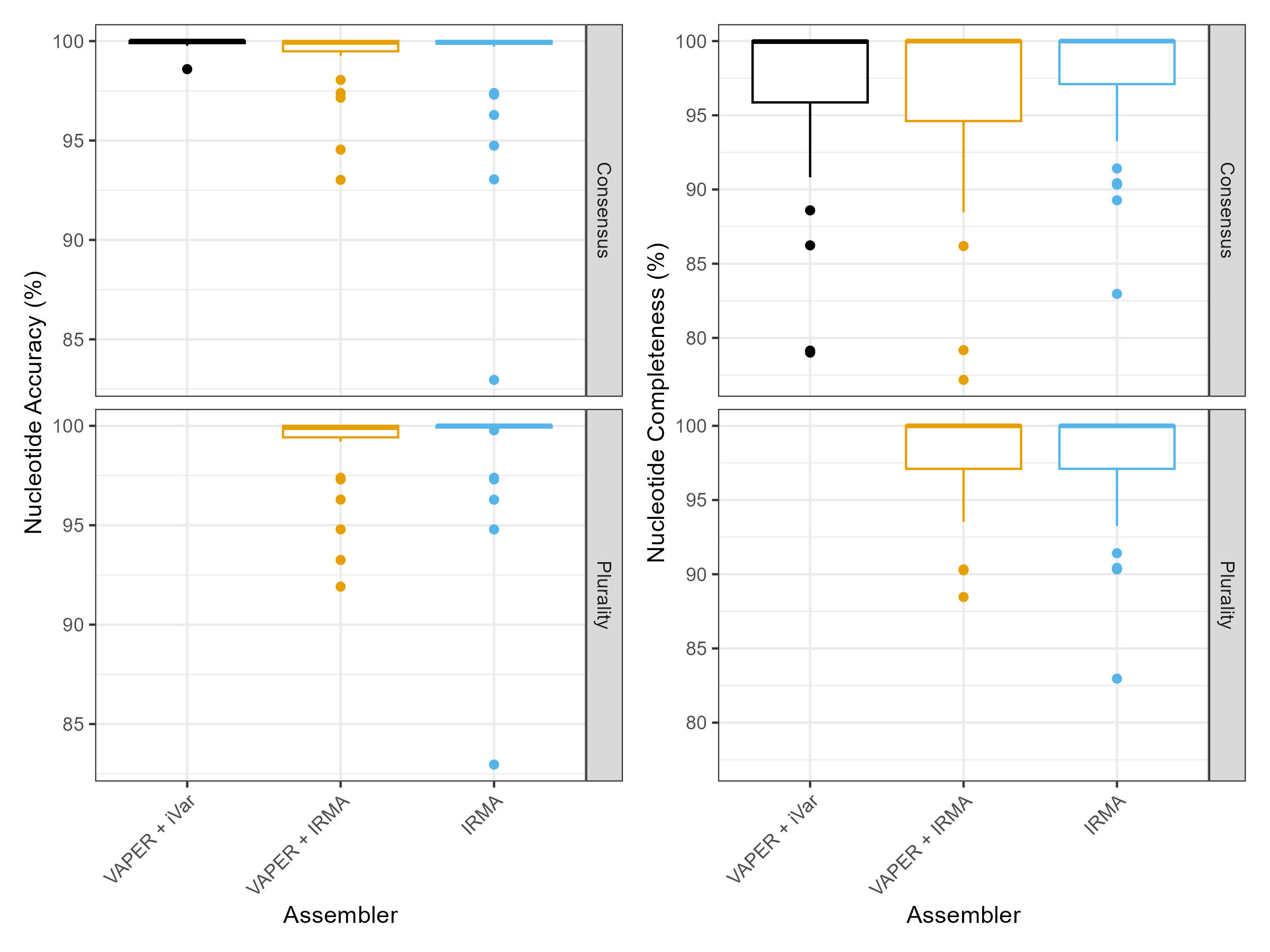
Assemblies were created from Twist Bioscience synthetic controls 4 and 17, using either VAPER and the EPITOME_2025-02-14 references or using the standard IRMA FLU module:
- VAPER + iVar:
--cons_assembler ivar --cons_assembly_type consensus - VAPER + IRMA (Consensus):
--cons_assembler irma --cons_assembly_type consensus - VAPER + IRMA (Plurality):
--cons_assembler irma --cons_assembly_type plurality - IRMA (Consensus / Plurality):
IRMA FLU --external-config <config> <read1> <read2> <outdir>
The minimum read length was adjusted to 75 bp for 3 of the 4 technical replicates of control 17, using the --external-config parameter when running the standard IRMA module.